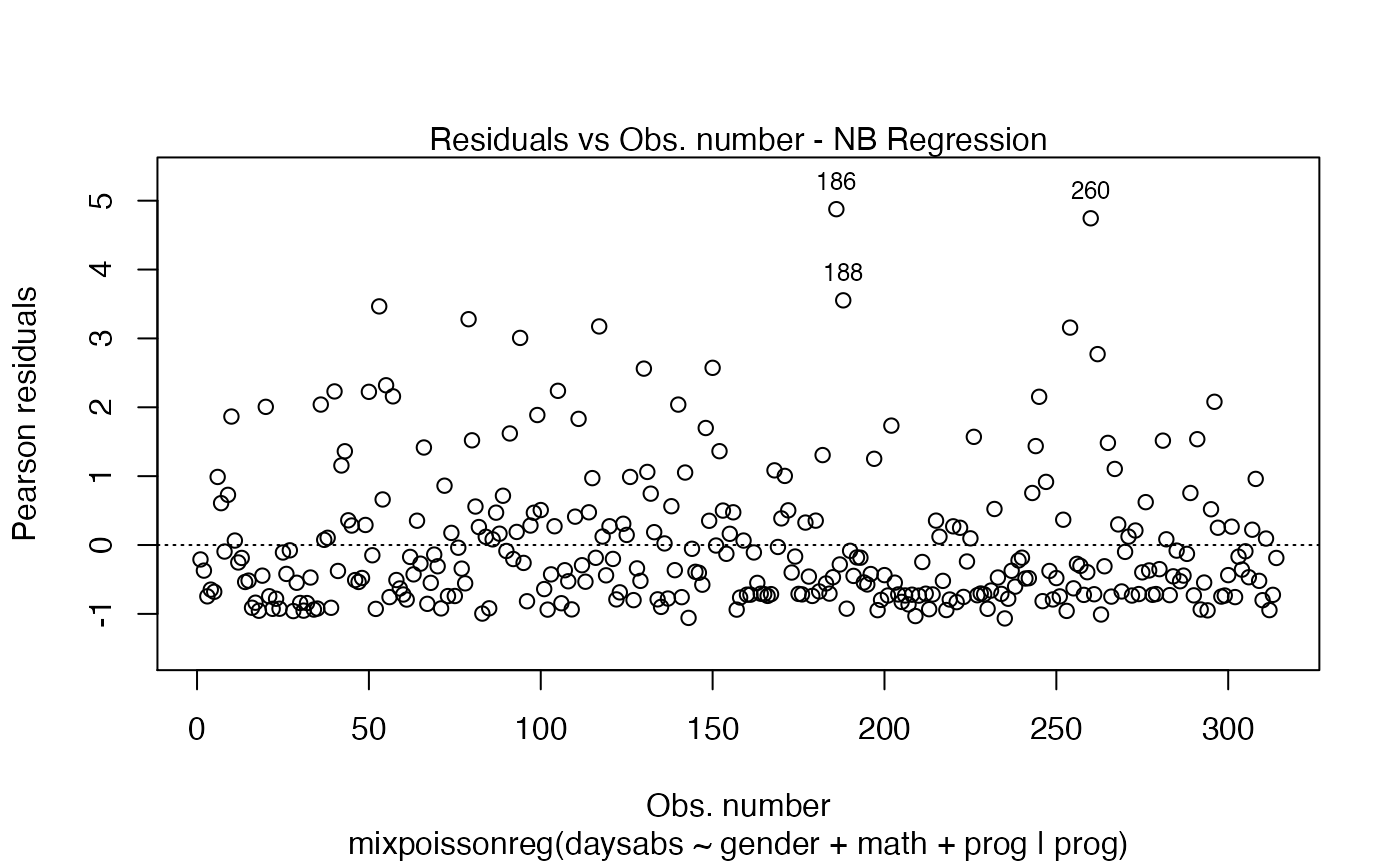
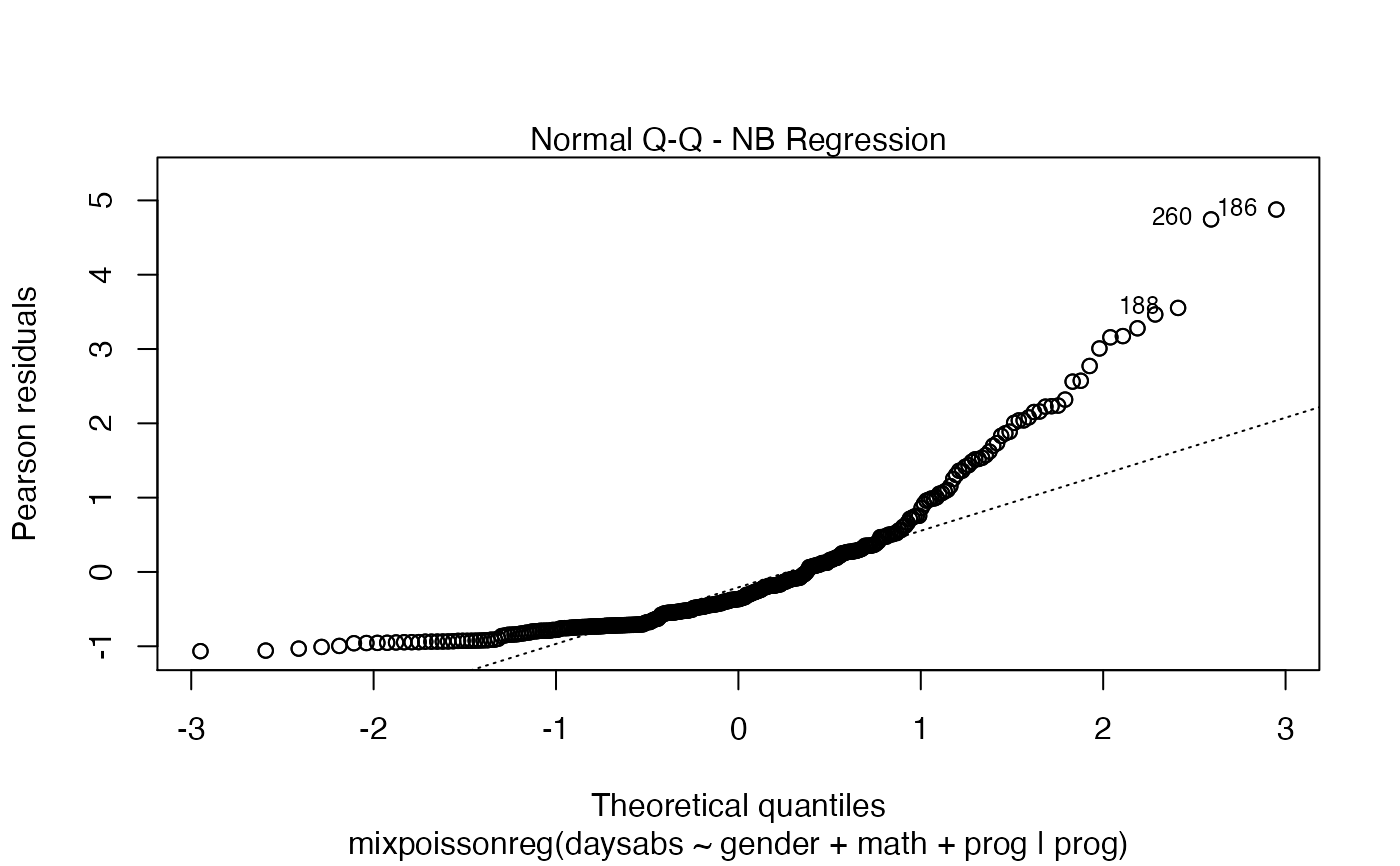
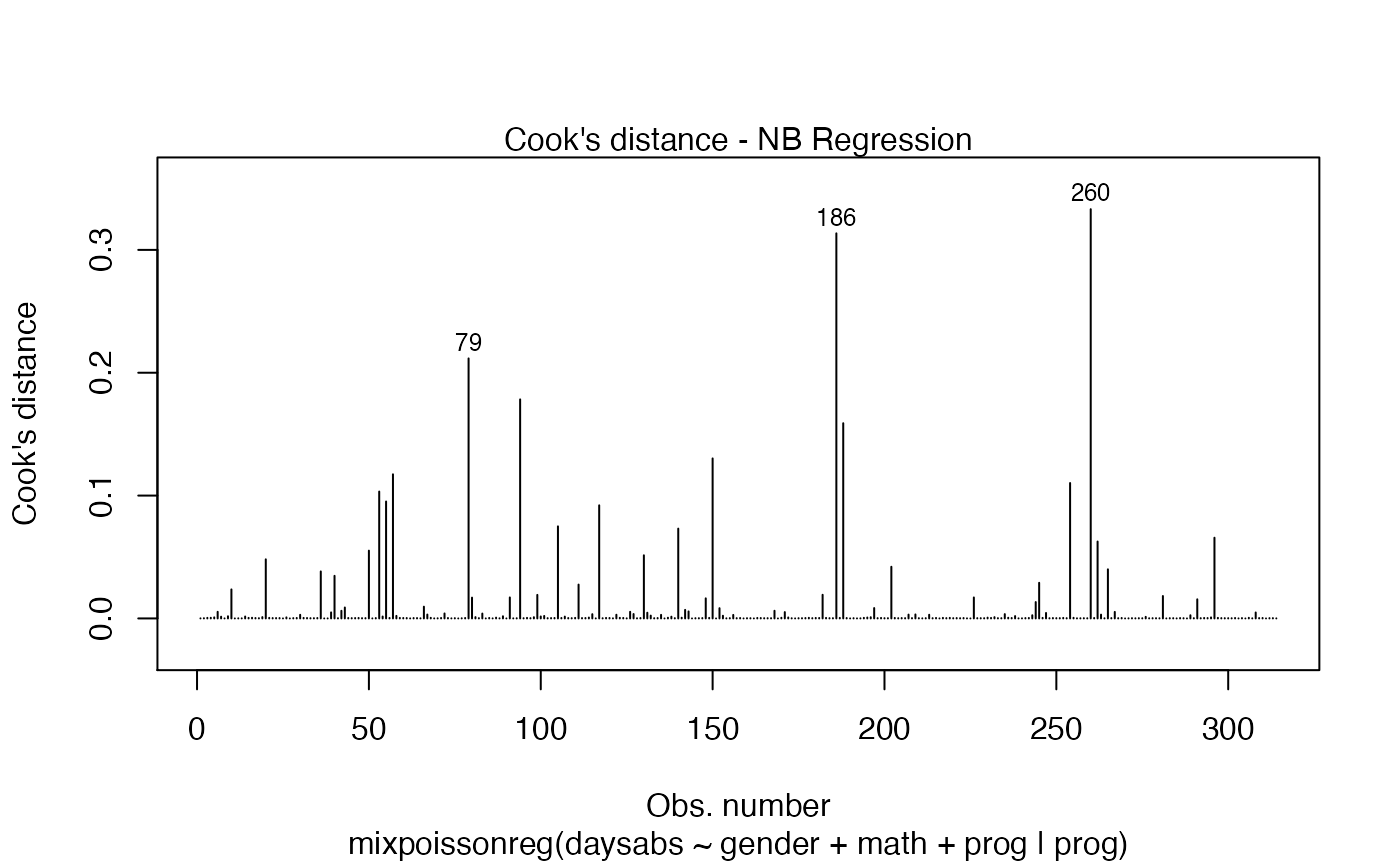
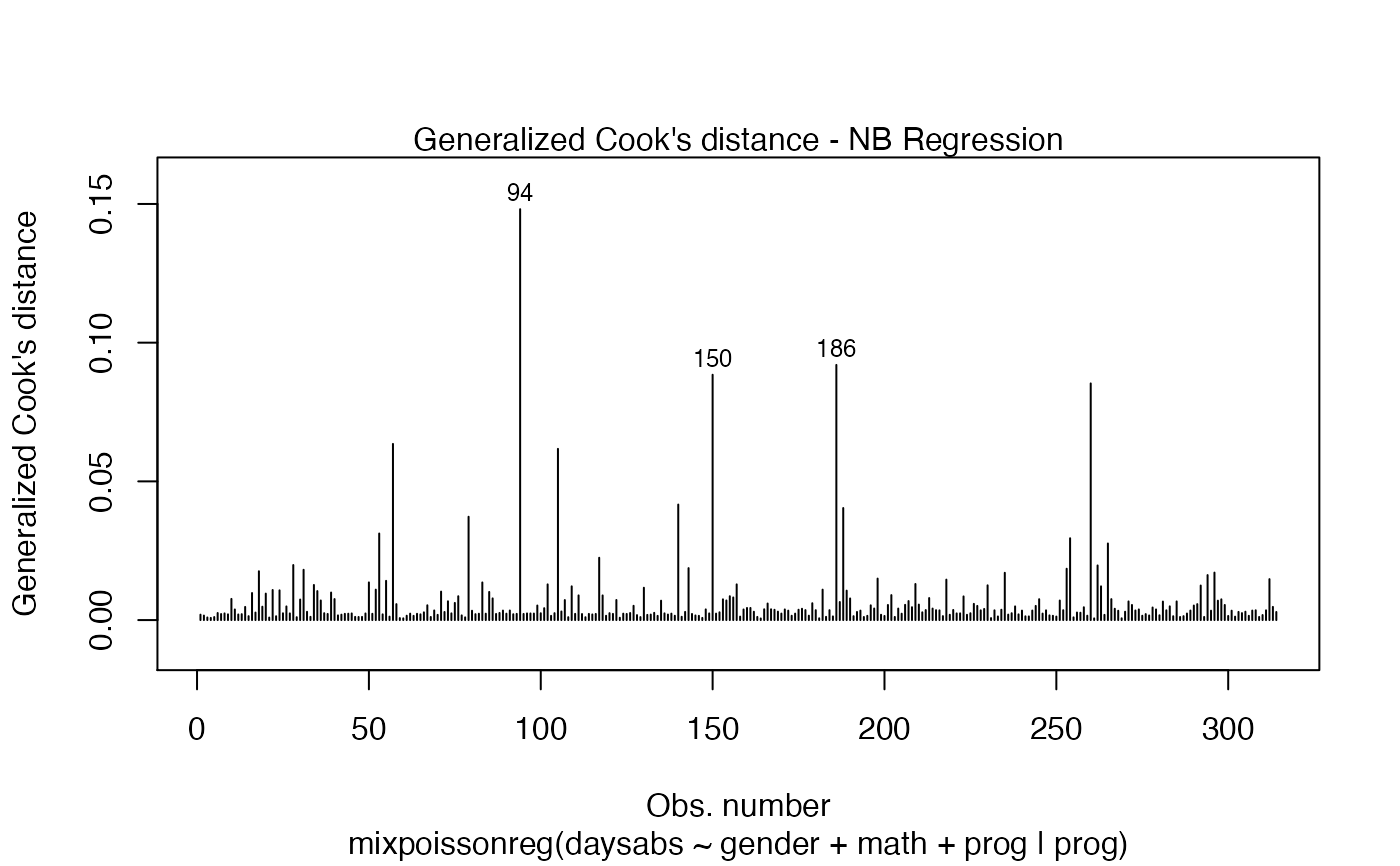
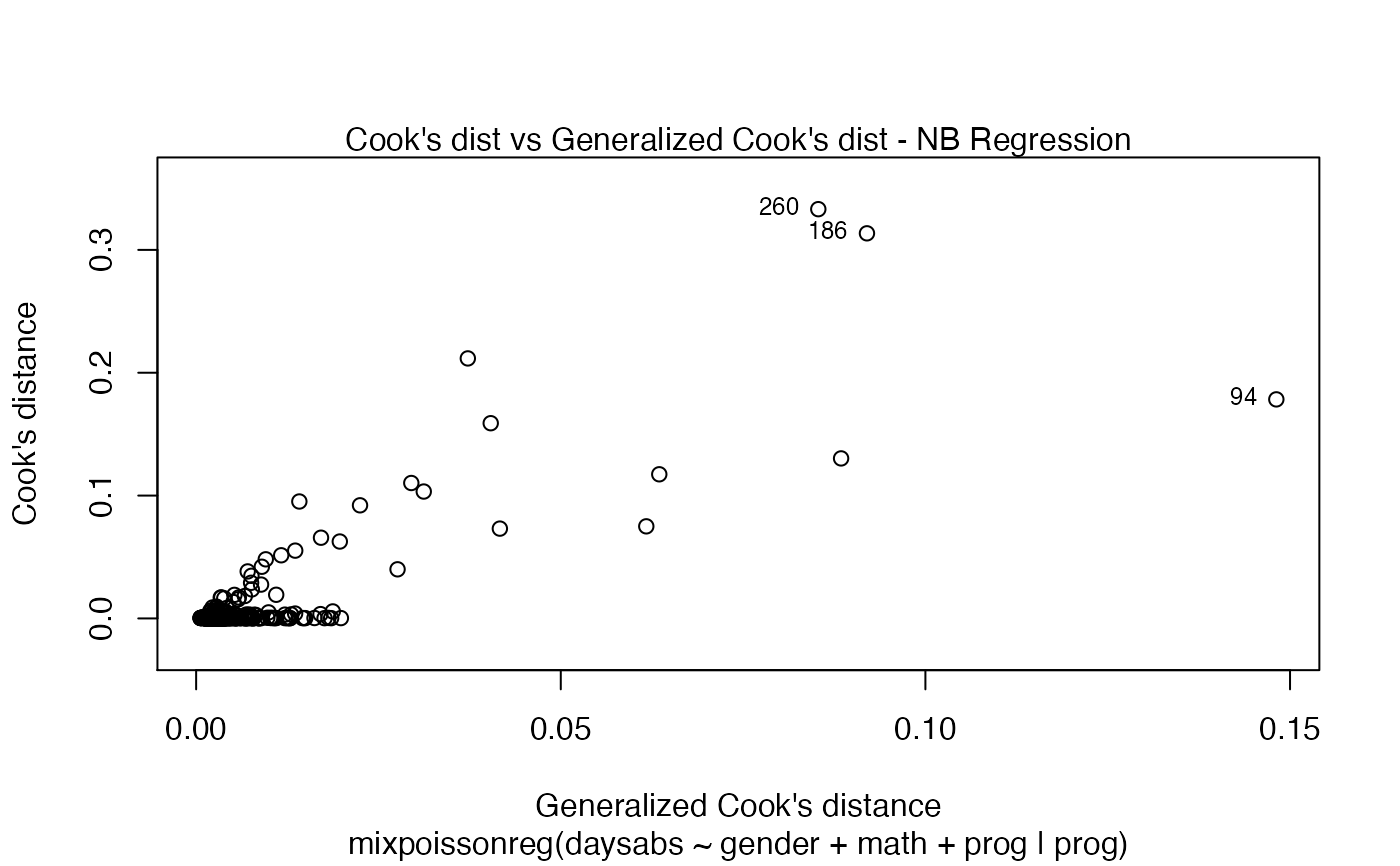
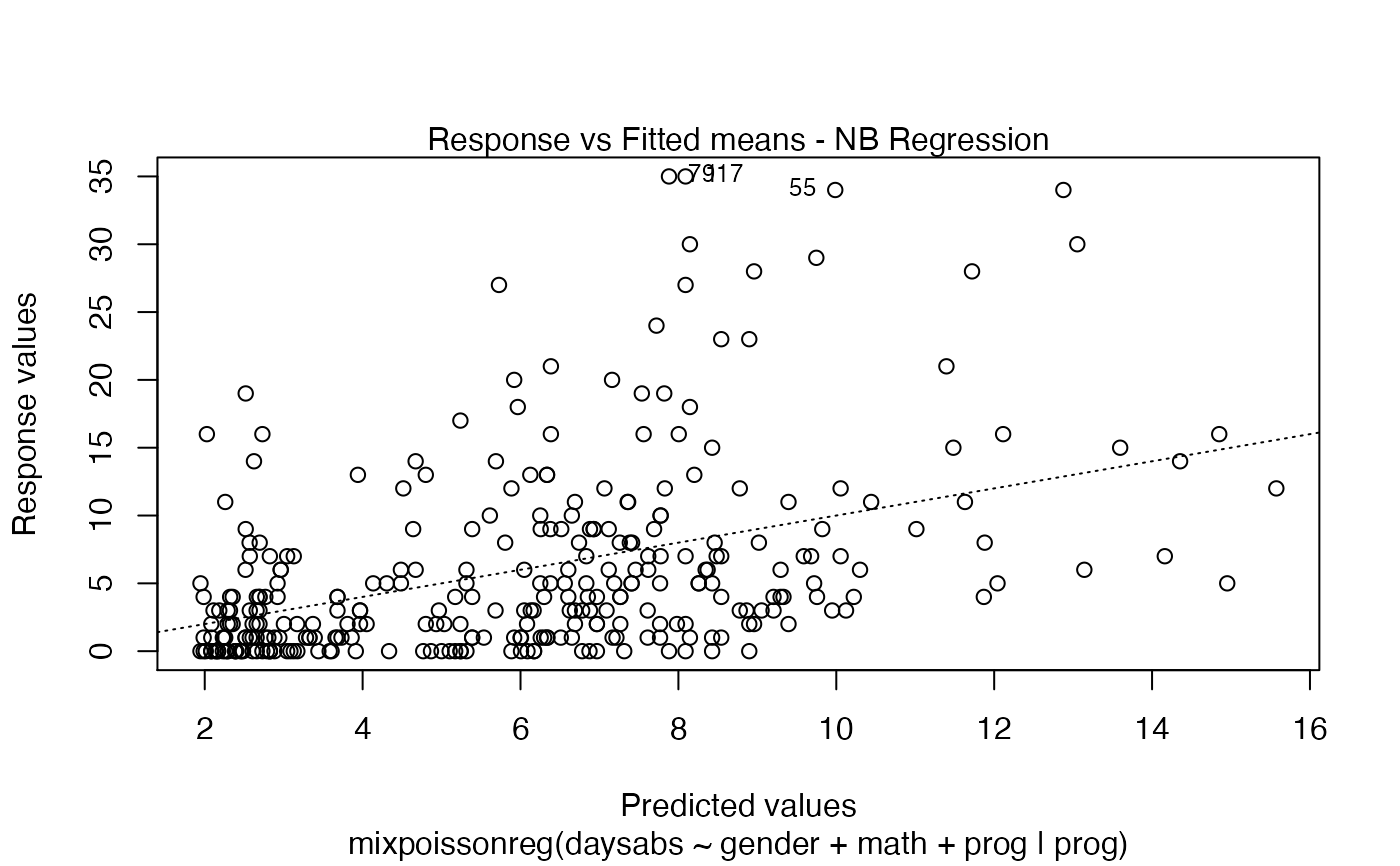
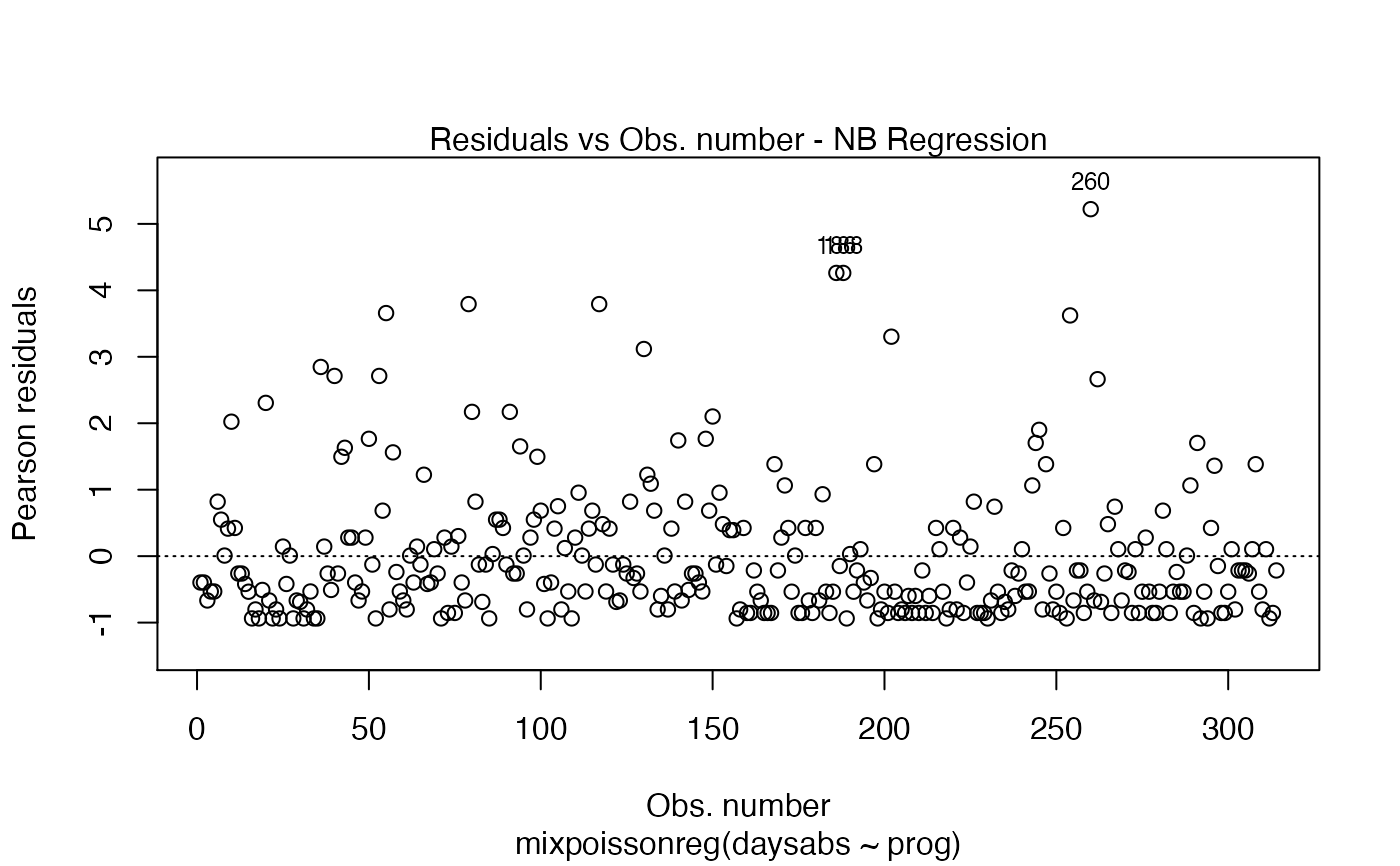
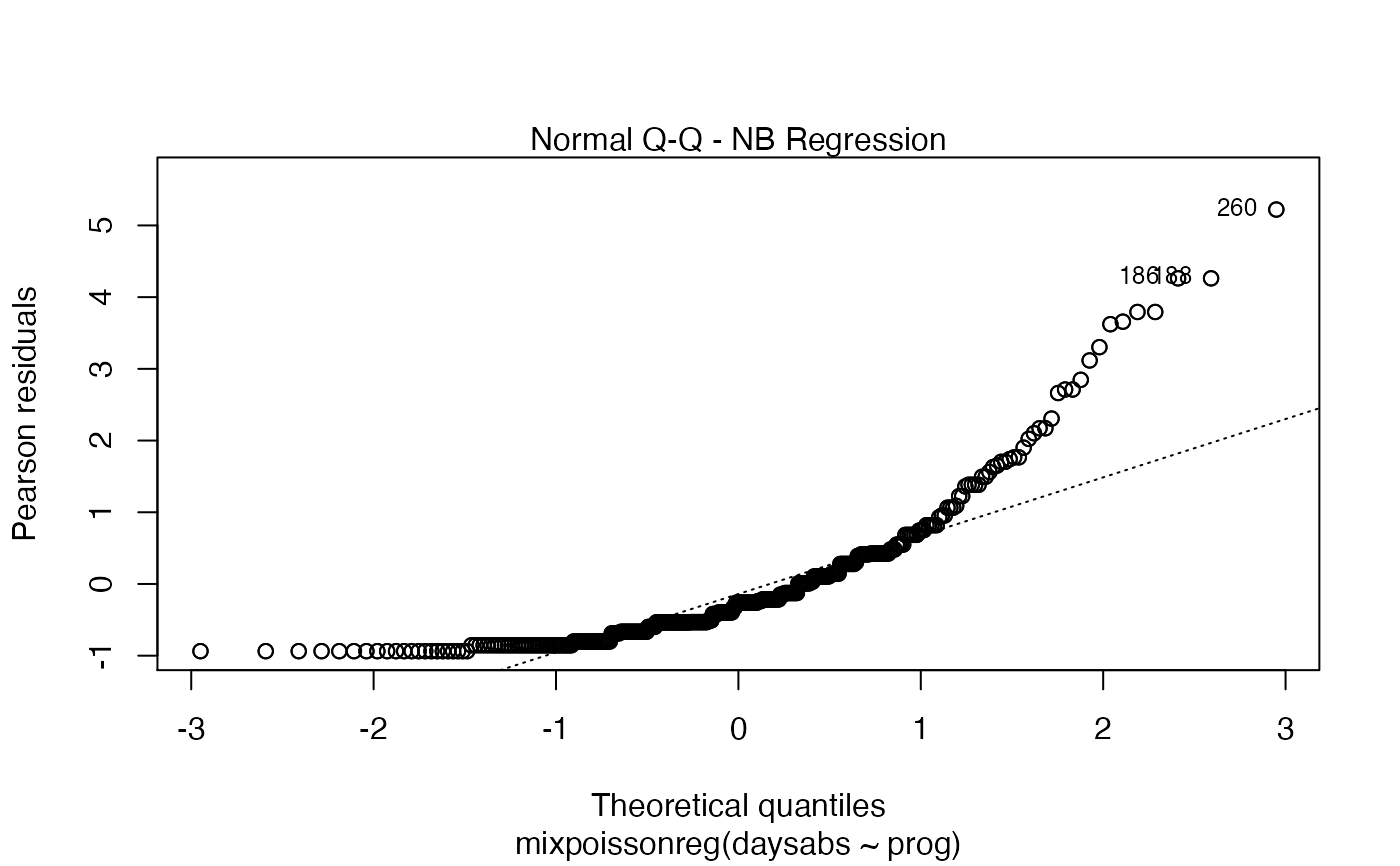
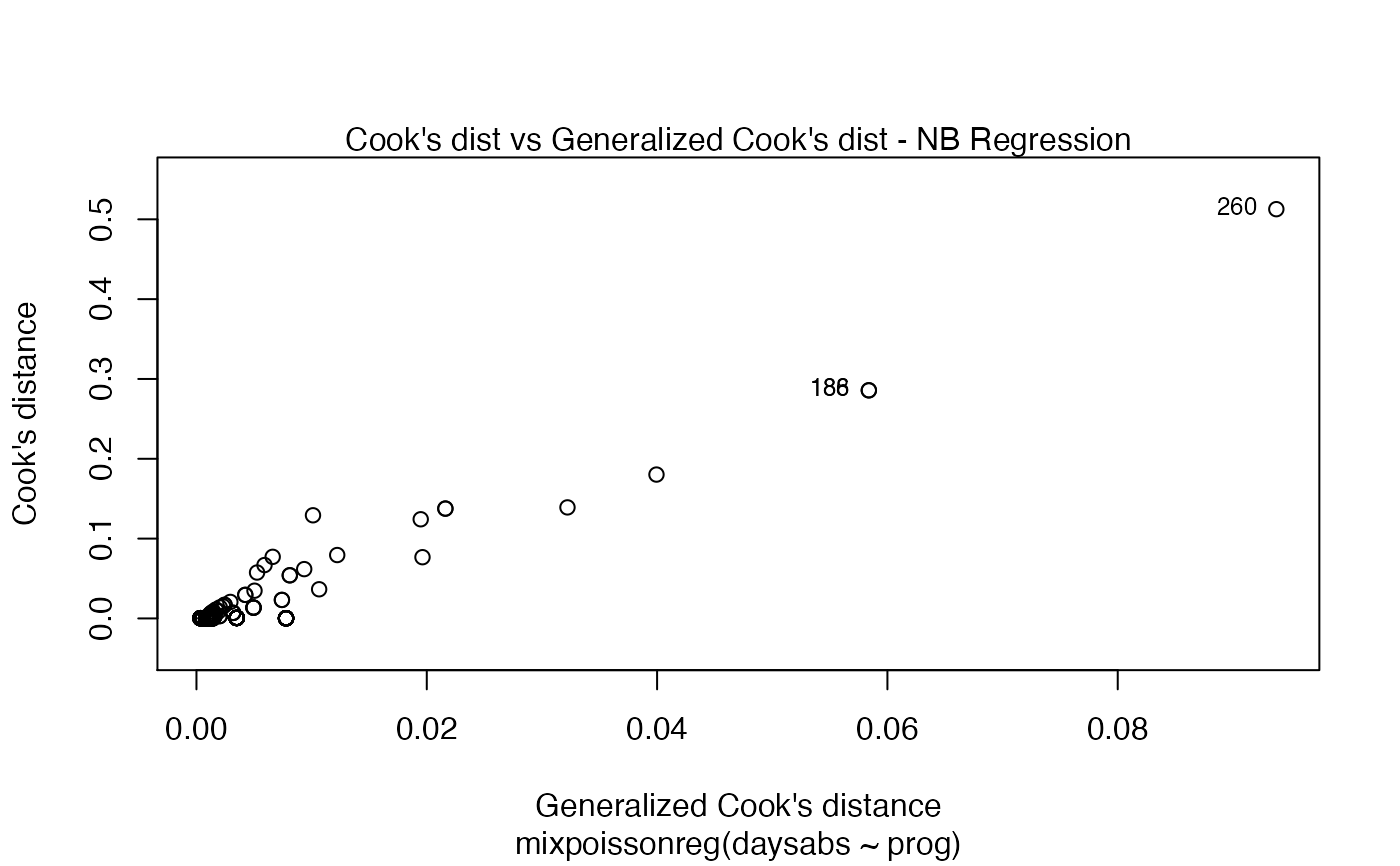
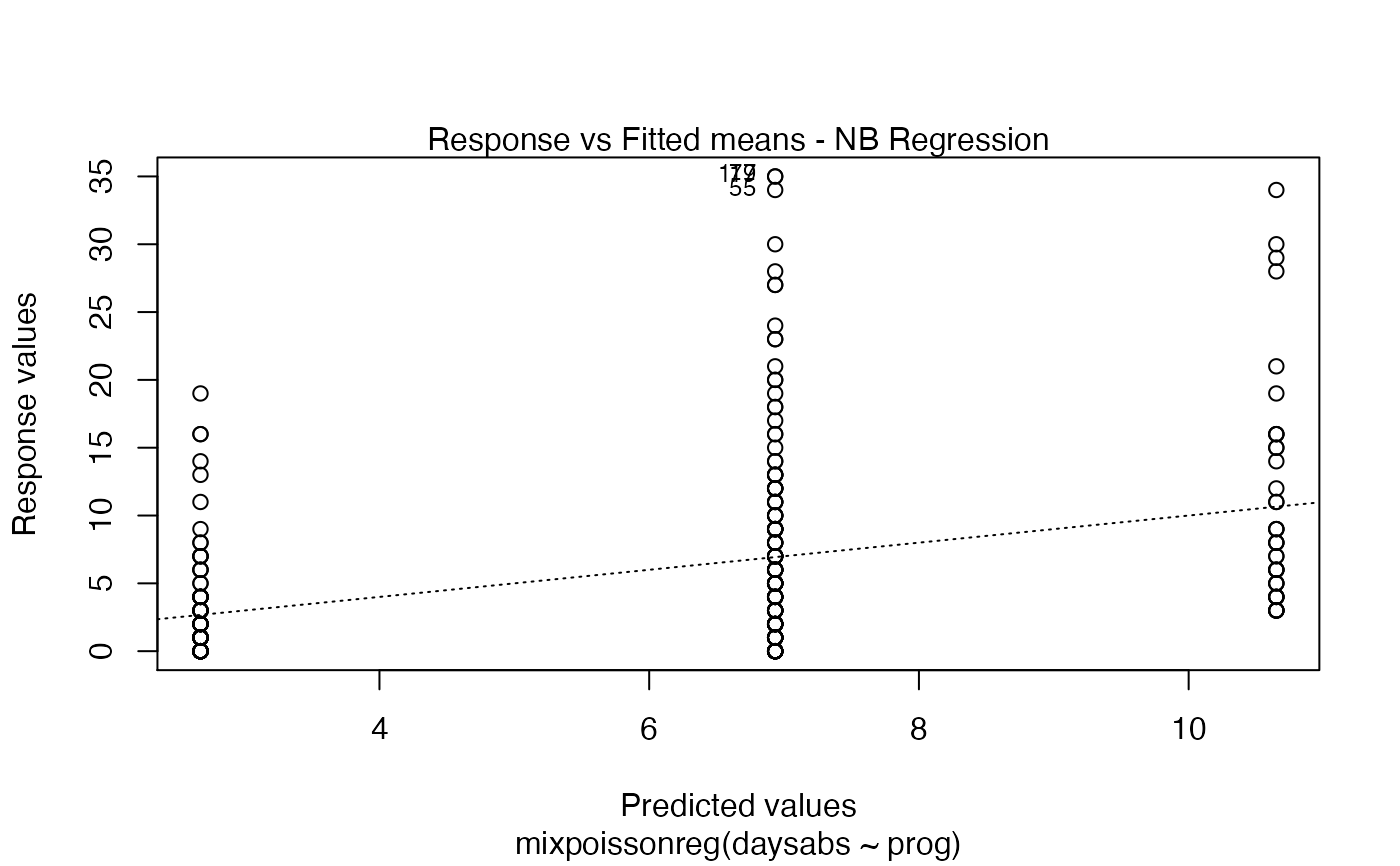
Currently there are six plots available. They contain residual analysis and global influence diagnostics. The plots are selectable by
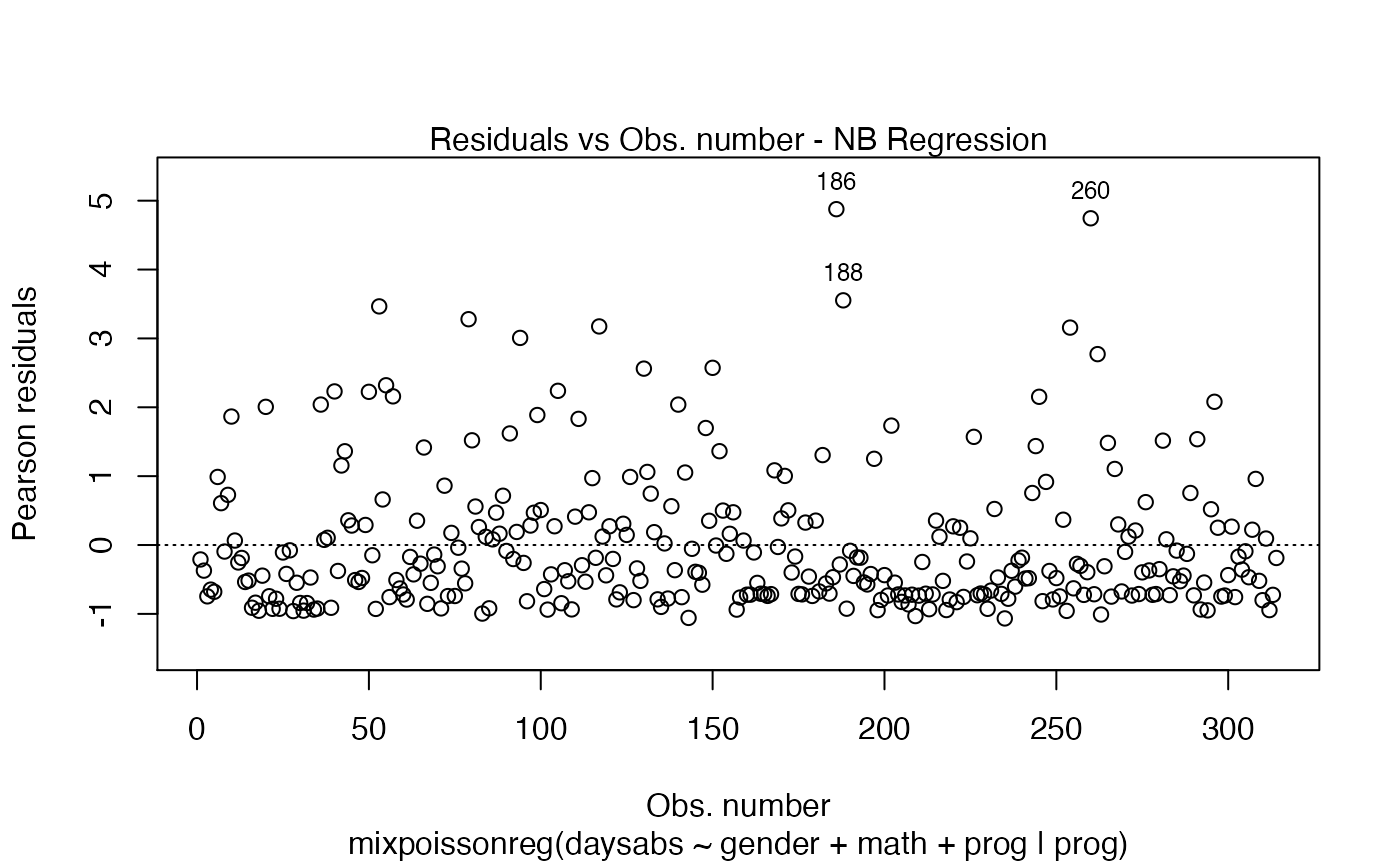
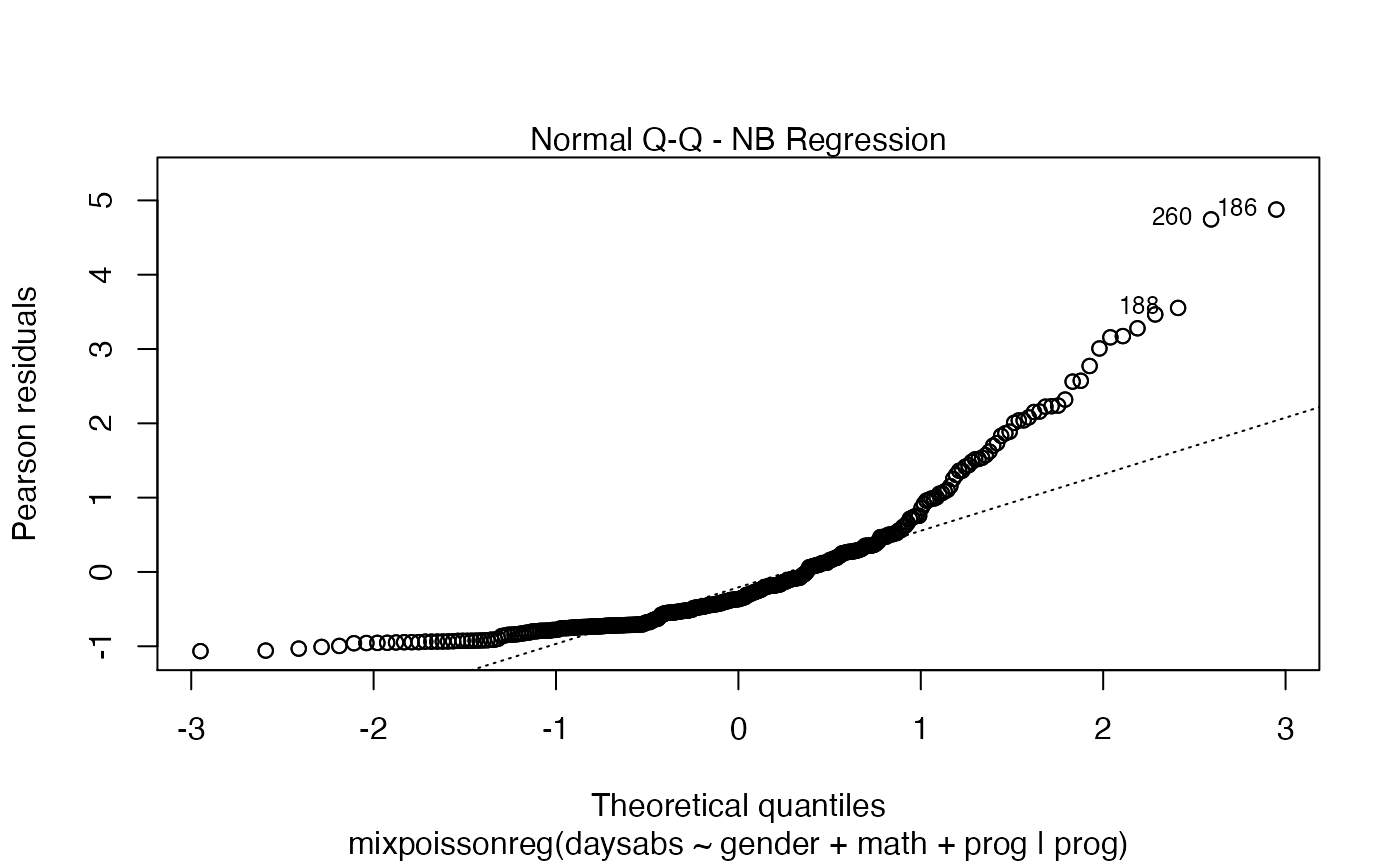
the which argument. The plots are: Residuals vs. obs. numbers; Normal Q-Q plots, which may contain simulated envelopes, if the fitted object
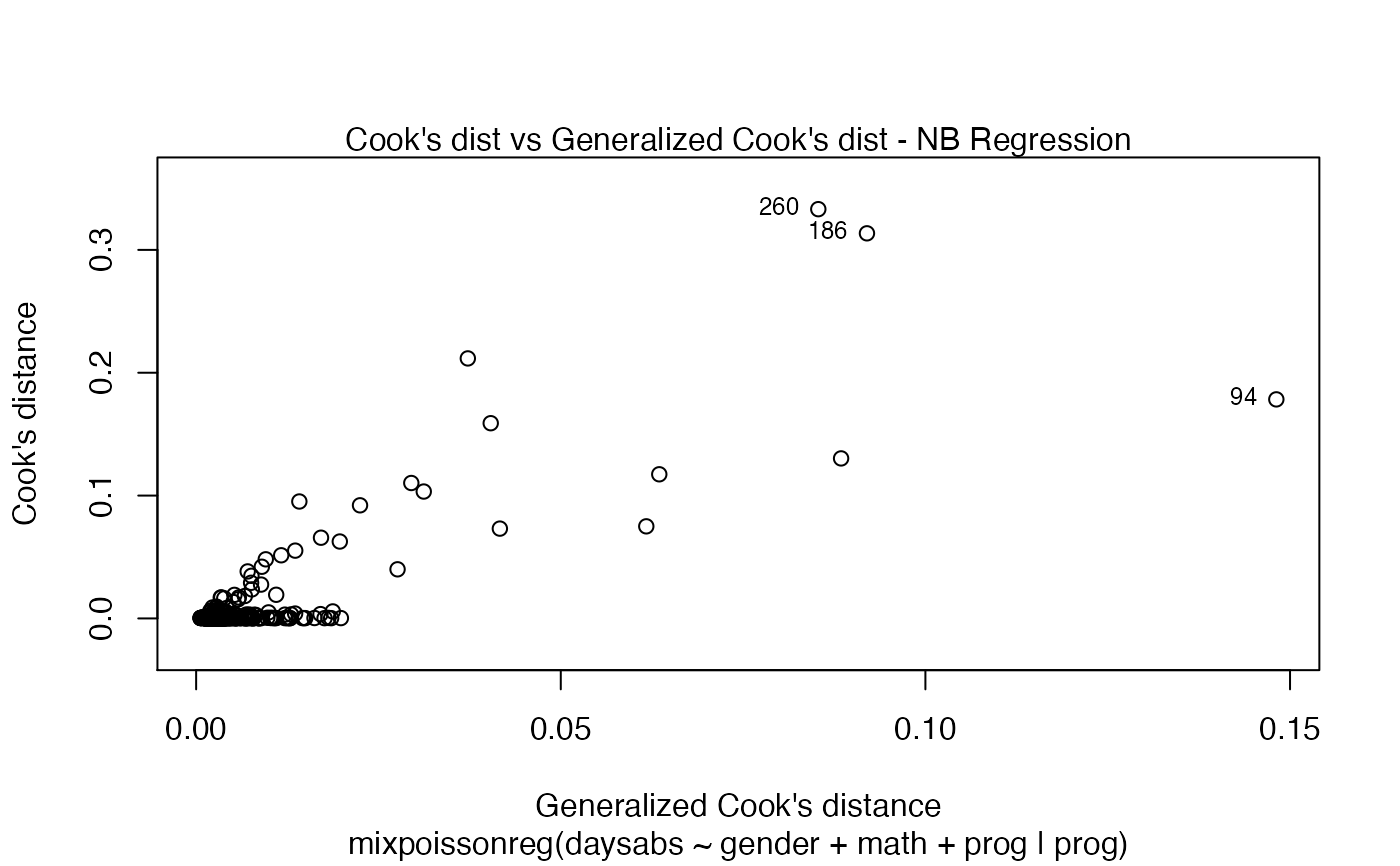
has simulated envelopes; Cook's distances vs. obs. numbers; Generalized Cook's distances vs. obs. numbers; Cook's distances vs. Generalized Cook's distances;
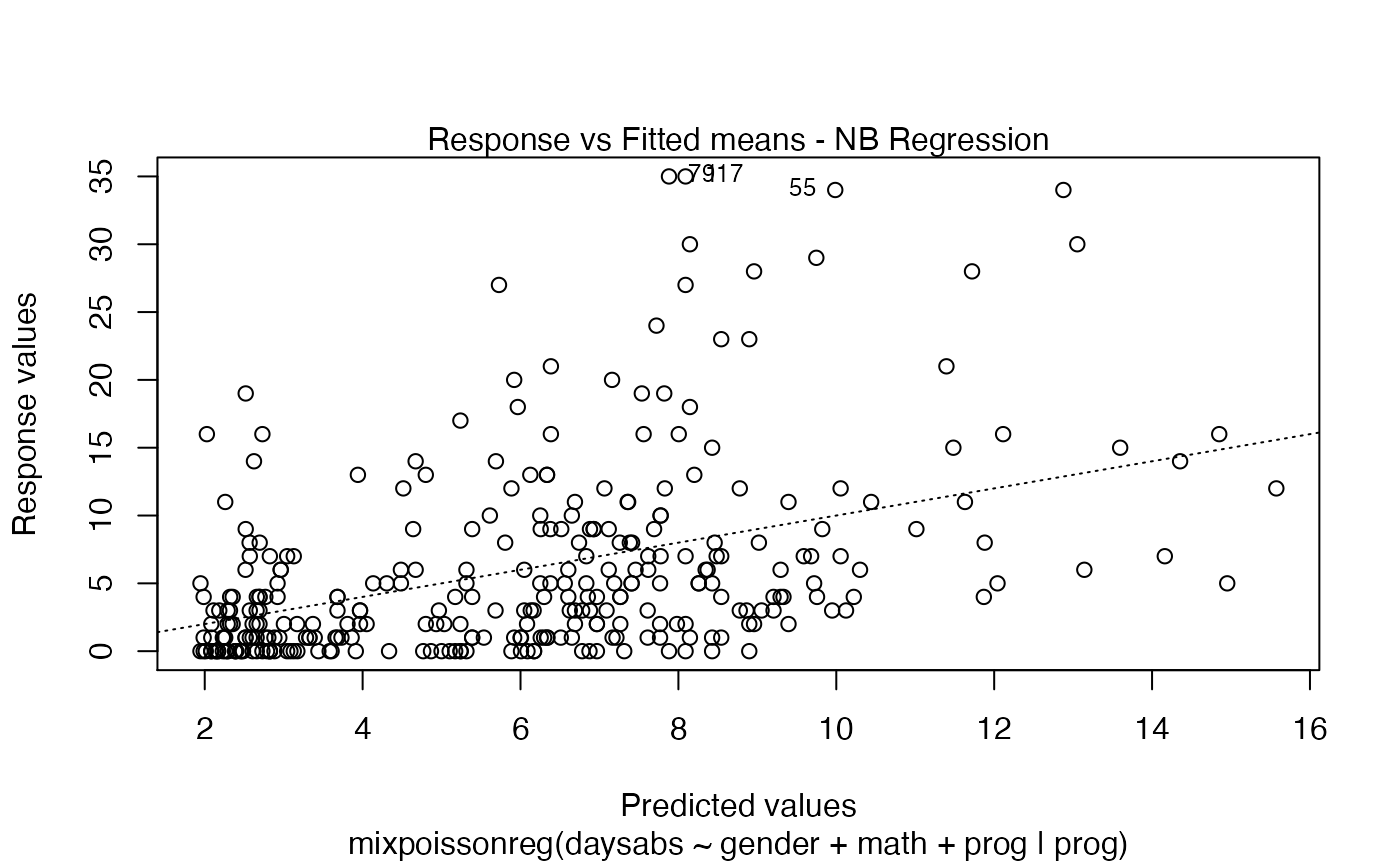
Response variables vs. fitted means. By default, the first two plots and the last two plots are provided.
# S3 method for mixpoissonreg plot( x, which = c(1, 2, 5, 6), caption = list("Residuals vs Obs. number", "Normal Q-Q", "Cook's distance", "Generalized Cook's distance", "Cook's dist vs Generalized Cook's dist", "Response vs Fitted means"), sub.caption = NULL, qqline = TRUE, col.qqline = "black", main = "", line_col_env = "gray", line_col_median = "black", fill_col_env = "gray", fill_alpha_env = 0.7, ask = prod(graphics::par("mfcol")) < length(which) && grDevices::dev.interactive(), labels.id = names(stats::residuals(x)), label.pos = c(4, 2), type.cookplot = "h", id.n = 3, col.id = NULL, cex.id = 0.75, cex.oma.main = 1.25, cex.caption = 1, col.caption = "black", cex.points = 1, col.points = "black", include.modeltype = TRUE, include.residualtype = FALSE, ... )
Arguments
| x | object of class "mixpoissonreg" containing results from the fitted model. If the model was fitted with envelope = 0, the Q-Q plot will be produced without envelopes. |
|---|---|
| which | a list or vector indicating which plots should be displayed. If a subset of the plots is required, specify a subset of the numbers 1:6,
see caption below for the different kinds. In
plot number 2, 'Normal Q-Q', if the |
| caption | captions to appear above the plots; character vector or list of valid graphics annotations. Can be set to "" or NA to suppress all captions. |
| sub.caption | common title-above the figures if there are more than one. If NULL, as by default, a possible abbreviated version of |
| qqline | logical; if |
| col.qqline | color of the qqline. |
| main | character; title to be placed at each plot additionally (and above) all captions. |
| line_col_env | line color for the upper and lower quantile curves of the simulated envelopes if the |
| line_col_median | line color for the median curve of the simulated envelopes if the |
| fill_col_env | fill color for the simulated envelopes if the |
| fill_alpha_env | alpha of the envelope region, when the |
| ask | logical; if |
| labels.id | vector of labels, from which the labels for extreme points will be chosen. The default uses the observation numbers. |
| label.pos | positioning of labels, for the left half and right half of the graph respectively, for plots 2 and 6. |
| type.cookplot | character; what type of plot should be drawn for Cook's and Generalized Cook's distances (plots 3 and 4). The default is 'h'. |
| id.n | number of points to be labelled in each plot, starting with the most extreme. |
| col.id | color of point labels. |
| cex.id | magnification of point labels. |
| cex.oma.main | controls the size of the sub.caption only if that is above the figures when there is more than one. |
| cex.caption | controls the size of caption. |
| col.caption | controls the caption color. |
| cex.points | controls the size of the points. |
| col.points | controls the colors of the points. |
| include.modeltype | logical. Indicates whether the model type ('NB' or 'PIG') should be displayed on the captions. |
| include.residualtype | local. Indicates whether the name of the residual ('Pearson' or 'Score') should be displayed on the caption of plot 1 (Residuals vs. Index). |
| ... | graphical parameters to be passed. |
Value
It is called for its side effects.
Details
The plot method is implemented following the same structure as the plot.lm, so it will be easy to be used by practitioners that
are familiar with glm objects.
These plots allows one to perform residuals analsysis and influence diagnostics. There are other global influence functions, see influence.mixpoissonreg.
See Barreto-Souza and Simas (2016), Cook and Weisberg (1982) and Zhu et al. (2001).
References
DOI:10.1007/s11222-015-9601-6 doi: 10.1007/s11222-015-9601-6 (Barreto-Souza and Simas; 2016)
Cook, D.R. and Weisberg, S. (1982) Residuals and Influence in Regression. (New York: Chapman and Hall, 1982)
Zhu, H.T., Lee, S.Y., Wei, B.C., Zhu, J. (2001) Case-deletion measures formodels with incomplete data. Biometrika, 88, 727–737.
See also
autoplot.mixpoissonreg, local_influence_plot.mixpoissonreg, local_influence_autoplot.mixpoissonreg,
summary.mixpoissonreg, predict.mixpoissonreg, influence.mixpoissonreg
Examples
# \donttest{ data("Attendance", package = "mixpoissonreg") daysabs_fit <- mixpoissonreg(daysabs ~ gender + math + prog | prog, data = Attendance) plot(daysabs_fit, which = 1:6)daysabs_fit_ml <- mixpoissonregML(daysabs ~ gender + math + prog | prog, data = Attendance, envelope = 100) plot(daysabs_fit_ml, which = 2)